From Tools to Systems: Why I'm Building bioArena

In the next two years, research labs will have access to hundreds of specialized AI models - for docking, enzyme design, retrosynthesis, immunogenicity, ADMET prediction, and beyond. Every major AI lab will claim their model is the best for science.
How will scientists choose? Right now, we read papers, trust marketing, and ask colleagues. But as models multiply, that approach won't scale. We'll need a way to see which AI systems actually work on problems that matter, validated by researchers who understand the domain.
bioArena is infrastructure for that future.
A platform where AI systems compete on real research problems, judged by scientists who understand the domain. Not just correctness, but the strategy, tool use, and reasoning quality.
Here's how I got here.
Rowan MCP: Building the Tool
When AlphaFold, Chai, and Boltz started rolling out, I was excited. These were game-changing models for structural biology, but as someone from bio, not tech, the friction was immediate. Installing dependencies, setting up environments, debugging errors - every step assumed I had backend engineering skills.
Around this time, I discovered Rowan. They already solved what I was struggling with by wrapping dozens of computational chemistry tools into a single, scientist-friendly interface. No installation hell, no environment conflicts - just ready-to-use tools.
As I explored their platform, I realized that these tools were packaged in a way AI agents could understand. An agent could read your research context, select the right tool, and solve problems without you needing to know which model to use.
What if scientists could access all of Rowan's tools just by asking questions in plain English?
So I built Rowan MCP - a Model Context Protocol interface bridging natural language and computational chemistry. Now a researcher could ask "What's the pKa of aspirin?" and the AI handles everything: selecting the tool, formatting parameters, parsing results, and returning a clear answer without requiring any code.
Natural language query → tool selection → results
Having built Rowan MCP with Claude as my coding agent, I continued using Claude to run queries through the tool - asking it chem/bio questions, watching it select tools, checking its outputs. But I wondered: was Claude the best model for scientific tool use, or would other models handle these tasks better?
In conversations with the Rowan team, Corin expressed interest in benchmarking LLMs at tool-use for chemical problem solving. This conversation is what led me to my next question - what SOTA models were best at solving scientific problems when given access to the right tools?
LabAgents: Learning to Build Evaluations
Answering "which model is best at scientific tool use" was more complicated than I thought. With no ML background, I figured I'd write a few test queries, run them through different models, and compare outputs - maybe an afternoon's work. Instead, I stumbled into a field I didn't know existed: evaluations (or as they say, "evals"). What started as a quick comparison script turned into a multi-week project called LabAgents.
I built the first version: 10 chemistry questions, binary scoring, Claude as the single judge. Quick and dirty, but it exposed problems I hadn't considered:
- Models giving correct answers without calling tools (defeating the purpose)
- Inconsistent and incomplete tracing
- Sole judge bias
- Binary scoring that loses nuance
So I iterated on LabAgents and addressed these gaps:
- 22 questions across 3 difficulty tiers
- Multi-dimensional scoring
- Web-search enabled judges
- Multiple judges
Claude models dominated at 85-88% weighted scores, while others ranged from 34-70%. But the most interesting finding was o3 - despite sophisticated reasoning capabilities, it scored only 33.7% because it submitted computational jobs then left the chat (literally) without waiting for results.
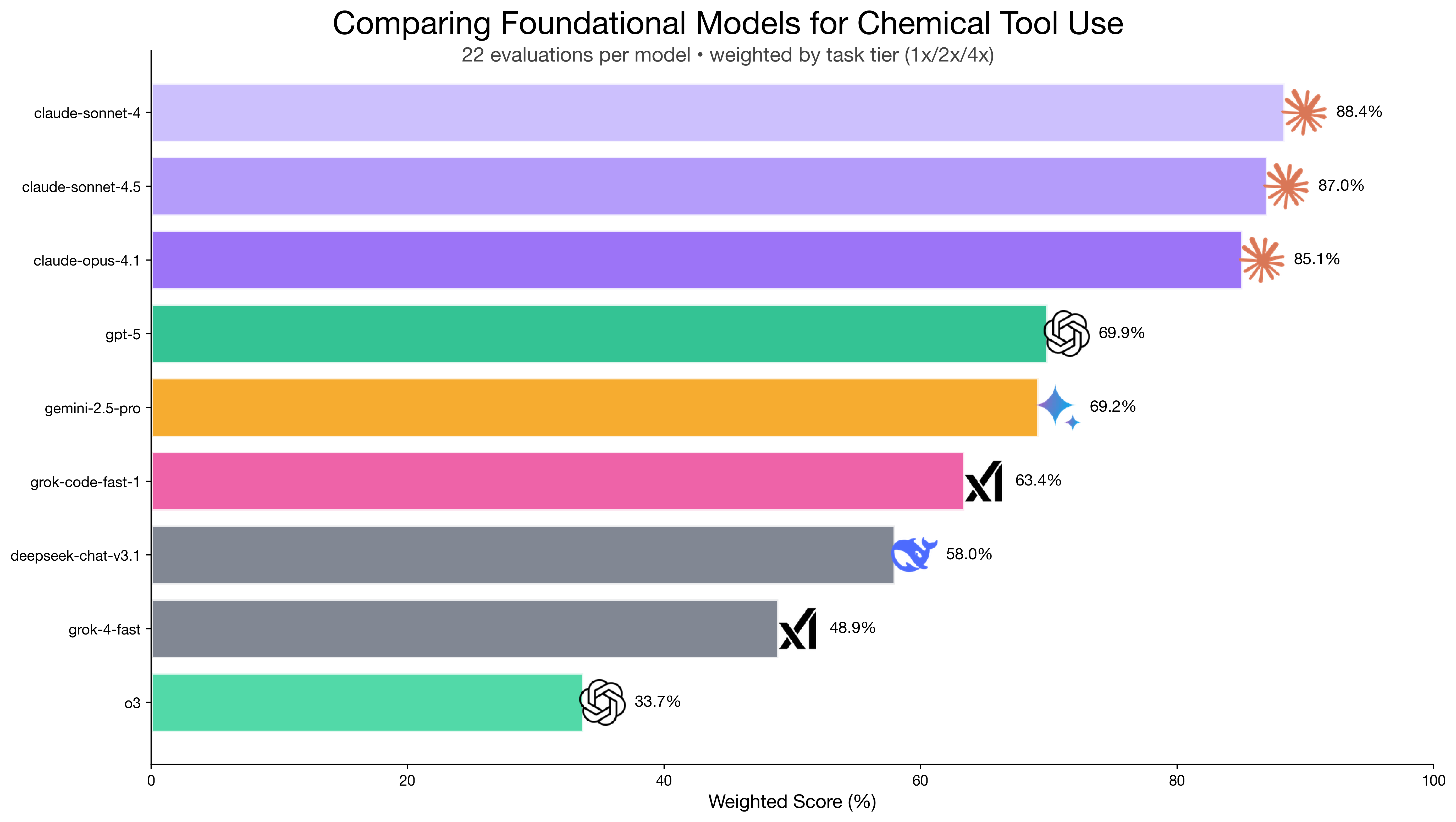
Claude models lead at 85-88%, while o3's 33.7% revealed that reasoning capability ≠ tool orchestration.
While these changes made evaluations more robust, I was still dealing with a core issue: without reasoning traces, meaningful evaluations are impossible. I need to know a scientist's strategy for solving a problem - tools and parameters they'd use, validation steps, and research between tool calls.
My solution: bioArena.
bioArena: Capturing How Science Gets Done
In a world where the intersection of science and AI continues to grow, we need to capture something we've never documented: reasoning traces that define scientific thinking. Not just the right answers, but the tool selection, hypothesis iteration, and the complex reality of research.
Current benchmarks are useful for validation, but have two limitations: (1) saturation as models improve, and (2) isolation from real workflows. Real research works differently. It demands complete systems where agents and tools orchestrate together to solve real-world multi-step problems.
So bioArena asks the question that matters: which combination of agents and tools actually works best for real scientific problems? Here, scientists and AI models tackle the same challenging problems side-by-side, documenting every decision - which tools they choose, the order of operations, where their reasoning forks. Domain experts then validate not just final answers, but the strategies used to get there.
This is integration testing for scientific AI - validating the entire stack, not just individual components. As agents evolve and new tools launch, bioArena adapts its evaluation framework. This ongoing assessment ensures you deploy solutions that advance your research today, not those that looked good on paper last month.
Come Build With Me
This journey has been built in the open - from Rowan MCP to LabAgents, and now bioArena. Science works best with transparency and collaboration. AI evaluation for science should too.
Whether you're a researcher exploring AI tools or a developer building for science, come contribute. Submit questions, validate approaches, or just follow along.
Check out Rowan MCP, the LabAgents repository, explore bioArena, or reach out on X or LinkedIn.

Thank you to Derek Alia and Corin Wagen for their feedback and guidance on earlier drafts of this post!